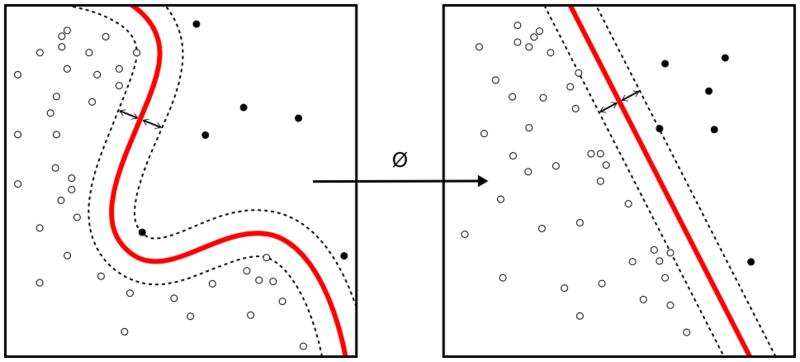
Support Vector Machine is one of the most commonly used supervised machine learning algorithms for data classification. A binary classifier, the support vector machine algorithm works in vector space to sort data points by finding the best hyperplane separating them into two groups. Thanks to its reliance upon vectors, it finds frontiers between groups of data points even in nonlinear patterns and features spaces of high dimensions.
To go into the details of support vector machine – often abbreviated SVM – and to program it in Python, this complete series of tutorials by Harrison Kinsley, a.k.a. Sentdex, dives in all the details of the support vector machine classifier, programming tricks and present example uses of the algorithm.
The following videos regroup the Support Vector Machine tutorials from the Machine Learning with Python series (parts 20 to 33) by Sentdex on Youtube.
The final code incorporating Kernels, a soft-margin Support Vector Machine, and Quadratic programming with CVXOPT can also be obtained from Sendex Python Programming website, recycling from an old (disappeared!) post from Mathieu Blondel.
For a complete presentation of this binary classifier, with mathematical formulas and extra developments, check the Wikipedia page on the Support Vector Machine algorithms.
To go further in each tutorial video, check the comments of the video on Youtube and the corresponding tutorial on the PythonProgramming website.
# Mathieu Blondel, September 2010
# License: BSD 3 clause
# http://www.mblondel.org/journal/2010/09/19/support-vector-machines-in-python/
# visualizing what translating to another dimension does
# and bringing back to 2D:
# https://www.youtube.com/watch?v=3liCbRZPrZA
# Docs: http://cvxopt.org/userguide/coneprog.html#quadratic-programming
# Docs qp example: http://cvxopt.org/examples/tutorial/qp.html
# Nice tutorial:
# https://courses.csail.mit.edu/6.867/wiki/images/a/a7/Qp-cvxopt.pdf
import numpy as np
from numpy import linalg
import cvxopt
import cvxopt.solvers
def linear_kernel(x1, x2):
return np.dot(x1, x2)
def polynomial_kernel(x, y, p=3):
return (1 + np.dot(x, y)) ** p
def gaussian_kernel(x, y, sigma=5.0):
return np.exp(-linalg.norm(x-y)**2 / (2 * (sigma ** 2)))
class SVM(object):
def __init__(self, kernel=linear_kernel, C=None):
self.kernel = kernel
self.C = C
if self.C is not None: self.C = float(self.C)
def fit(self, X, y):
n_samples, n_features = X.shape
# Gram matrix
K = np.zeros((n_samples, n_samples))
for i in range(n_samples):
for j in range(n_samples):
K[i,j] = self.kernel(X[i], X[j])
P = cvxopt.matrix(np.outer(y,y) * K)
q = cvxopt.matrix(np.ones(n_samples) * -1)
A = cvxopt.matrix(y, (1,n_samples))
b = cvxopt.matrix(0.0)
if self.C is None:
G = cvxopt.matrix(np.diag(np.ones(n_samples) * -1))
h = cvxopt.matrix(np.zeros(n_samples))
else:
tmp1 = np.diag(np.ones(n_samples) * -1)
tmp2 = np.identity(n_samples)
G = cvxopt.matrix(np.vstack((tmp1, tmp2)))
tmp1 = np.zeros(n_samples)
tmp2 = np.ones(n_samples) * self.C
h = cvxopt.matrix(np.hstack((tmp1, tmp2)))
# solve QP problem
solution = cvxopt.solvers.qp(P, q, G, h, A, b)
# Lagrange multipliers
a = np.ravel(solution['x'])
# Support vectors have non zero lagrange multipliers
sv = a > 1e-5
ind = np.arange(len(a))[sv]
self.a = a[sv]
self.sv = X[sv]
self.sv_y = y[sv]
print("%d support vectors out of %d points" % (len(self.a), n_samples))
# Intercept
self.b = 0
for n in range(len(self.a)):
self.b += self.sv_y[n]
self.b -= np.sum(self.a * self.sv_y * K[ind[n],sv])
self.b /= len(self.a)
# Weight vector
if self.kernel == linear_kernel:
self.w = np.zeros(n_features)
for n in range(len(self.a)):
self.w += self.a[n] * self.sv_y[n] * self.sv[n]
else:
self.w = None
def project(self, X):
if self.w is not None:
return np.dot(X, self.w) + self.b
else:
y_predict = np.zeros(len(X))
for i in range(len(X)):
s = 0
for a, sv_y, sv in zip(self.a, self.sv_y, self.sv):
s += a * sv_y * self.kernel(X[i], sv)
y_predict[i] = s
return y_predict + self.b
def predict(self, X):
return np.sign(self.project(X))
if __name__ == "__main__":
import pylab as pl
def gen_lin_separable_data():
# generate training data in the 2-d case
mean1 = np.array([0, 2])
mean2 = np.array([2, 0])
cov = np.array([[0.8, 0.6], [0.6, 0.8]])
X1 = np.random.multivariate_normal(mean1, cov, 100)
y1 = np.ones(len(X1))
X2 = np.random.multivariate_normal(mean2, cov, 100)
y2 = np.ones(len(X2)) * -1
return X1, y1, X2, y2
def gen_non_lin_separable_data():
mean1 = [-1, 2]
mean2 = [1, -1]
mean3 = [4, -4]
mean4 = [-4, 4]
cov = [[1.0,0.8], [0.8, 1.0]]
X1 = np.random.multivariate_normal(mean1, cov, 50)
X1 = np.vstack((X1, np.random.multivariate_normal(mean3, cov, 50)))
y1 = np.ones(len(X1))
X2 = np.random.multivariate_normal(mean2, cov, 50)
X2 = np.vstack((X2, np.random.multivariate_normal(mean4, cov, 50)))
y2 = np.ones(len(X2)) * -1
return X1, y1, X2, y2
def gen_lin_separable_overlap_data():
# generate training data in the 2-d case
mean1 = np.array([0, 2])
mean2 = np.array([2, 0])
cov = np.array([[1.5, 1.0], [1.0, 1.5]])
X1 = np.random.multivariate_normal(mean1, cov, 100)
y1 = np.ones(len(X1))
X2 = np.random.multivariate_normal(mean2, cov, 100)
y2 = np.ones(len(X2)) * -1
return X1, y1, X2, y2
def split_train(X1, y1, X2, y2):
X1_train = X1[:90]
y1_train = y1[:90]
X2_train = X2[:90]
y2_train = y2[:90]
X_train = np.vstack((X1_train, X2_train))
y_train = np.hstack((y1_train, y2_train))
return X_train, y_train
def split_test(X1, y1, X2, y2):
X1_test = X1[90:]
y1_test = y1[90:]
X2_test = X2[90:]
y2_test = y2[90:]
X_test = np.vstack((X1_test, X2_test))
y_test = np.hstack((y1_test, y2_test))
return X_test, y_test
def plot_margin(X1_train, X2_train, clf):
def f(x, w, b, c=0):
# given x, return y such that [x,y] in on the line
# w.x + b = c
return (-w[0] * x - b + c) / w[1]
pl.plot(X1_train[:,0], X1_train[:,1], "ro")
pl.plot(X2_train[:,0], X2_train[:,1], "bo")
pl.scatter(clf.sv[:,0], clf.sv[:,1], s=100, c="g")
# w.x + b = 0
a0 = -4; a1 = f(a0, clf.w, clf.b)
b0 = 4; b1 = f(b0, clf.w, clf.b)
pl.plot([a0,b0], [a1,b1], "k")
# w.x + b = 1
a0 = -4; a1 = f(a0, clf.w, clf.b, 1)
b0 = 4; b1 = f(b0, clf.w, clf.b, 1)
pl.plot([a0,b0], [a1,b1], "k--")
# w.x + b = -1
a0 = -4; a1 = f(a0, clf.w, clf.b, -1)
b0 = 4; b1 = f(b0, clf.w, clf.b, -1)
pl.plot([a0,b0], [a1,b1], "k--")
pl.axis("tight")
pl.show()
def plot_contour(X1_train, X2_train, clf):
pl.plot(X1_train[:,0], X1_train[:,1], "ro")
pl.plot(X2_train[:,0], X2_train[:,1], "bo")
pl.scatter(clf.sv[:,0], clf.sv[:,1], s=100, c="g")
X1, X2 = np.meshgrid(np.linspace(-6,6,50), np.linspace(-6,6,50))
X = np.array([[x1, x2] for x1, x2 in zip(np.ravel(X1), np.ravel(X2))])
Z = clf.project(X).reshape(X1.shape)
pl.contour(X1, X2, Z, [0.0], colors='k', linewidths=1, origin='lower')
pl.contour(X1, X2, Z + 1, [0.0], colors='grey', linewidths=1, origin='lower')
pl.contour(X1, X2, Z - 1, [0.0], colors='grey', linewidths=1, origin='lower')
pl.axis("tight")
pl.show()
def test_linear():
X1, y1, X2, y2 = gen_lin_separable_data()
X_train, y_train = split_train(X1, y1, X2, y2)
X_test, y_test = split_test(X1, y1, X2, y2)
clf = SVM()
clf.fit(X_train, y_train)
y_predict = clf.predict(X_test)
correct = np.sum(y_predict == y_test)
print("%d out of %d predictions correct" % (correct, len(y_predict)))
plot_margin(X_train[y_train==1], X_train[y_train==-1], clf)
def test_non_linear():
X1, y1, X2, y2 = gen_non_lin_separable_data()
X_train, y_train = split_train(X1, y1, X2, y2)
X_test, y_test = split_test(X1, y1, X2, y2)
clf = SVM(polynomial_kernel)
clf.fit(X_train, y_train)
y_predict = clf.predict(X_test)
correct = np.sum(y_predict == y_test)
print("%d out of %d predictions correct" % (correct, len(y_predict)))
plot_contour(X_train[y_train==1], X_train[y_train==-1], clf)
def test_soft():
X1, y1, X2, y2 = gen_lin_separable_overlap_data()
X_train, y_train = split_train(X1, y1, X2, y2)
X_test, y_test = split_test(X1, y1, X2, y2)
clf = SVM(C=1000.1)
clf.fit(X_train, y_train)
y_predict = clf.predict(X_test)
correct = np.sum(y_predict == y_test)
print("%d out of %d predictions correct" % (correct, len(y_predict)))
plot_contour(X_train[y_train==1], X_train[y_train==-1], clf)
#test_linear()
#test_non_linear()
test_soft()